My Research Projects
Elucidating the bacterial silver resistance
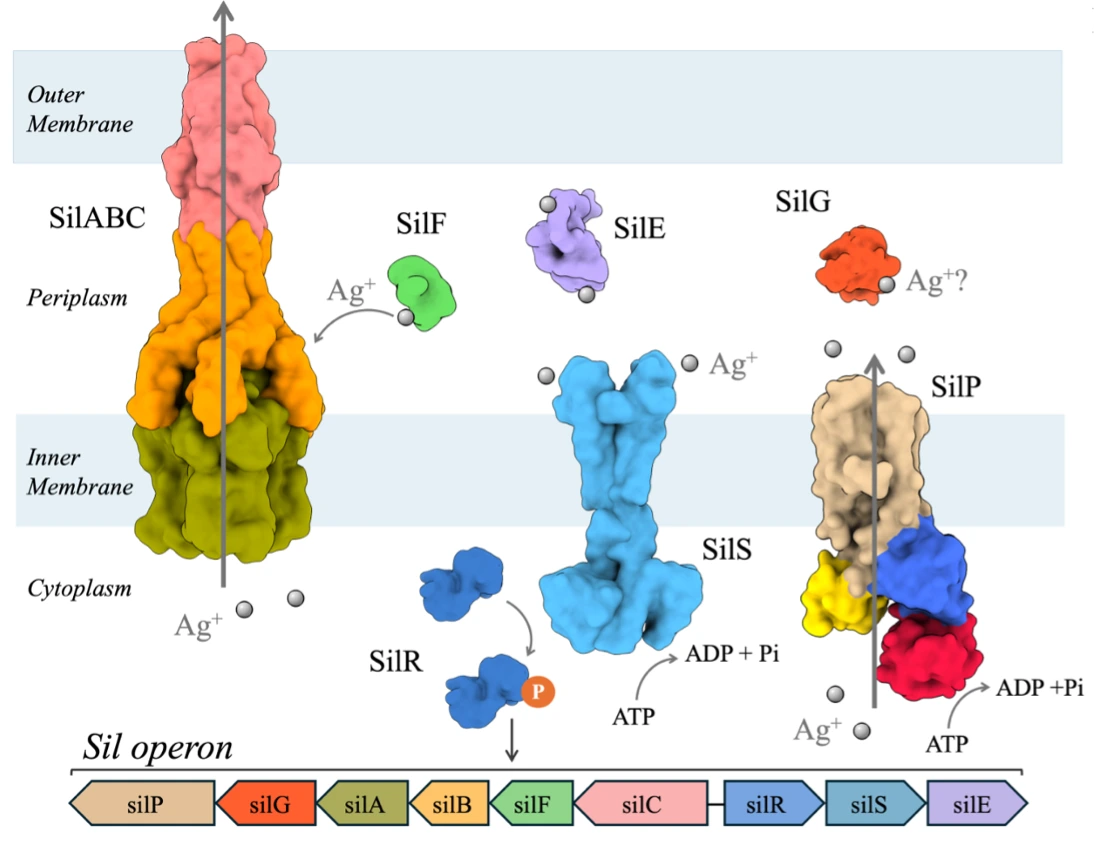
Silver antimicrobial properties have been used for thousands of years, from the Chaldeans as early as 4000 B.C.E. Despite this long-standing history and its demonstrated activity against Gram-negative bacteria, the complete bactericidal mode of action of silver remains unclear. To counteract the toxic effect of silver, Gram-negative bacteria have developed different resistance mechanisms, including the efficient transport of the metal out of the cell promoted by an efflux pump. The latter one is encoded by the silver-resistant gene cluster named sil composed of 9 proteins: an Ag+/H+ tripartite efflux pump (SilABC), a P-type ATPase efflux pump (SilP), a two-component regulatory system (SilRS) and three periplasmic silver-binding proteins SilE, SilG and SilF. First, our group investigated the structures of SilE mimicking peptides upon silver binding, then we provided structural and dynamical insights into full-length Ag+-bound SilE from combined analytical probes. Our group also made a step forward with the structural and dynamical characterization of the SilF/SilB complex, where they unraveled the mechanism by which SilF transfers silver ions and interacts with SilB that belongs to the efflux pump SilABC.
The Prediction of NMR spin relaxation by Molecular dynamics
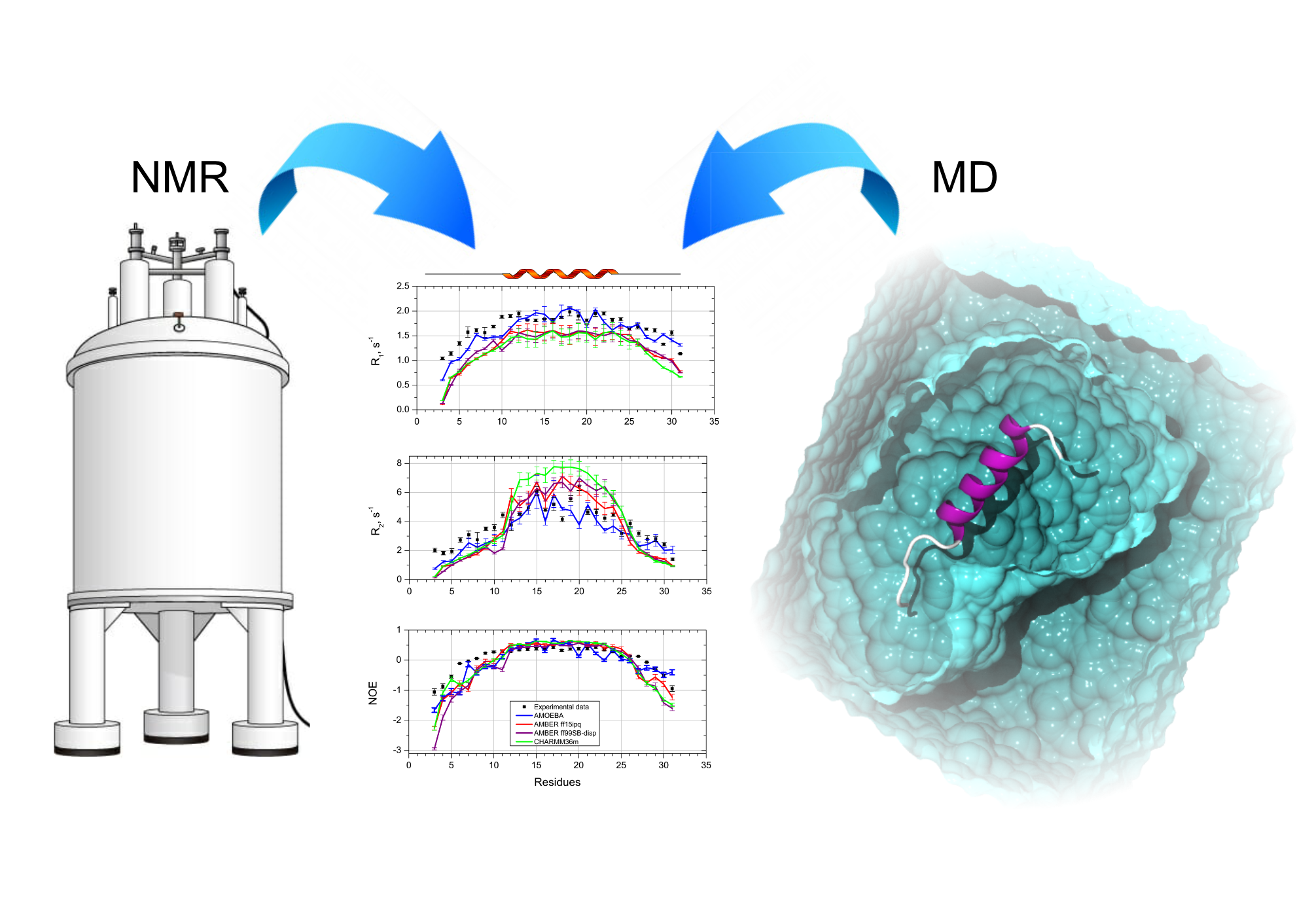
Dynamics is essential for proteins to perform interactions with biological partners. Among the vast landscape of biophysical methods, NMR possesses this exquisite potential to sample motions at different frequency through the measurement of various relaxation parameters. The interpretation of relaxation data is commonly subjected to the choice of a suitable model, like the model free approach, for instance. We are seeking to develop an approach based on molecular dynamics to predict all the available NMR spin relaxation. See references related to the use of quaternion to predict the rotational diffusion tensor, the prediction of R1, R2 and NOEs and the use of polarizable force fields.